BATCure
- Home
- The Project
- Project Goals
- Project Workplan
- Project Management Structure
- The Partners
- Dissemination
- Events
- Links
- Family Area
- Public Deliverables
- Contact us
Project Workplan
BATCure consists of a demanding plan of interconnected work over a period of 36 months designed to provide focus and maximum benefit and reach the end goal in the most effective and synergistic manner possible. The plan of work is divided into 8 experimental research Work Packages (WP1-8) and 2 supporting WPs (WP9,10). Each WP addresses a single objective and is divided into focused aims and tasks. All WPs except one (WP6) start at month 1 of the project. They work in parallel, closely linked to each other.
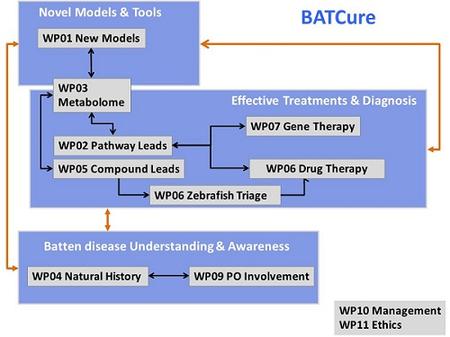
WP1 - New Models
Development, validation and optimisation of essential new models and tools
In this work package, new cell and small vertebrate models will be derived, or obtained, in order to supplement existing models for CLN3, CLN6 and CLN7 diseases and provide the most appropriate tools for developing and testing therapies.
WP2 - Pathway Leads
Identifying therapeutic target pathways using genetics and biology
The goal of this work package is to delineate disease biology and gene function as a means to identifying new therapeutic target pathways. This highly integrated WP will take three unbiased and three hypothesis-driven approaches in yeast and mammalian cell disease models to identify or confirm new target pathways using genetics, cell biology, metabolic, proteomic and bioinformatic approaches.
WP3 - Metabolome
Identifying therapeutic target pathways, and developing new diagnostic and monitoring techniques using metabonomics
In this WP, advanced metabolomic profiling techniques will be used for comprehensive characterisation of samples to compare healthy metabolomes with disease in order to i) identify disturbed biochemical pathways, ii) design new treatments with the objective of normalising the disturbed biochemical pathways, iii) identify the most appropriate models of each disease for evaluation of new chemical entities and iv) develop new diagnostic and monitoring techniques for other WPs.
WP4 - Natural History
Extending natural history beyond the brain
The focus of this WP is CLN3 disease, extending to CLN6 and CLN7 diseases by using relevant mouse models and zebrafish disease models to understand the cardiac defects.
WP5 - Compound Leads
Identifying new therapeutic compound leads
Multiparametric cell-based high content assays using disease relevant cell types and readouts for NCLs will be developed and validated. These assays will be instrumental to screen target focus libraries of many thousands of compounds (new and FDA-approved drugs) in order to identify correctors of NCL disease phenotypes.
WP6 - Zebrafish Triage
Determination of therapeutic potential using zebrafish
Novel zebrafish models from WP1 will be validated for hallmarks of NCL before new potentially disease modifying compounds identified in WP5 will be tested on the zebrafish disease models for their effect on lysosomal storage and/or epilepsy, motor skills and survival in a high-throughput assay.
WP7 - Gene Therapy
Gene therapy for the brain, eye and other organs
The main goal of this WP is to develop gene therapy protocols for the treatment of central and peripheral pathology in mouse models of CLN3, CLN6 and/or CLN7 deficiency.
WP8 - Drug Therapy
Delivering new small molecule therapy to the mouse (Month 1 to Month 36)
This WP aims at evaluating therapeutic potential of selected lead compounds in mouse models of CLN3, CLN6 and CLN7 diseases by use of immunohistochemical, neurochemical, and behavioural methods.
WP9 - Patient Organisation Involvement
This WP will be focused on i) developing a methodology to enable patients, patient groups and stakeholders to participate in the project; ii) preparing for future clinical trials; iii) devising a communication strategy with public and professional engagement; and iv)developing an exploitation and dissemination strategy.
WP10 Management
This Work Package incorporates duties covering all aspects of project monitoring, reporting, financial and contractual administration, ensuring proper communication within the consortium and implementing the project’s governance and decision-making structures.


